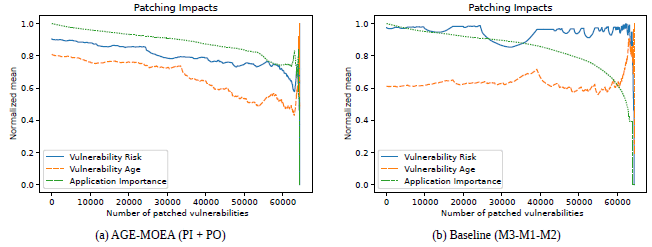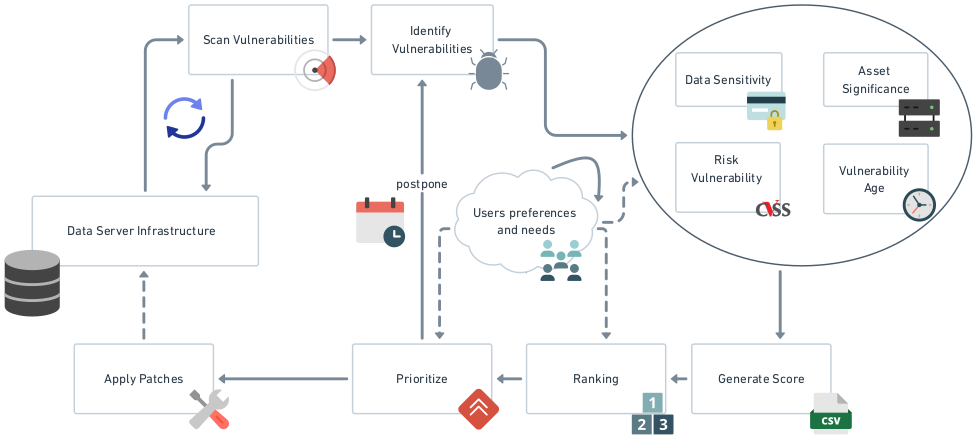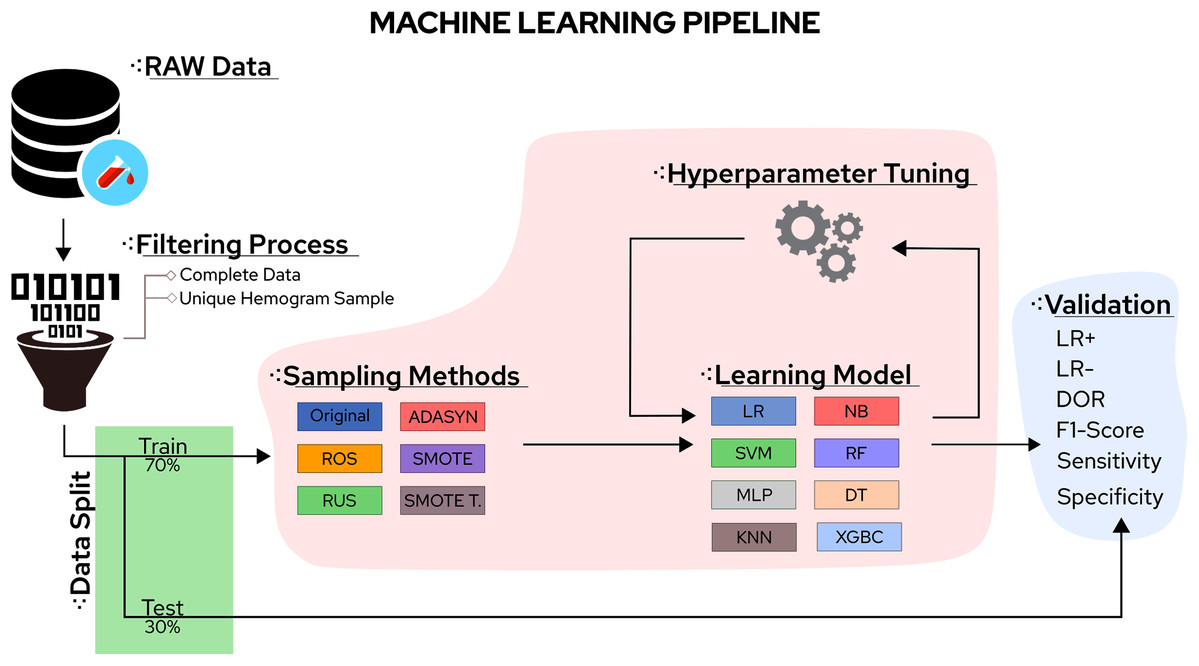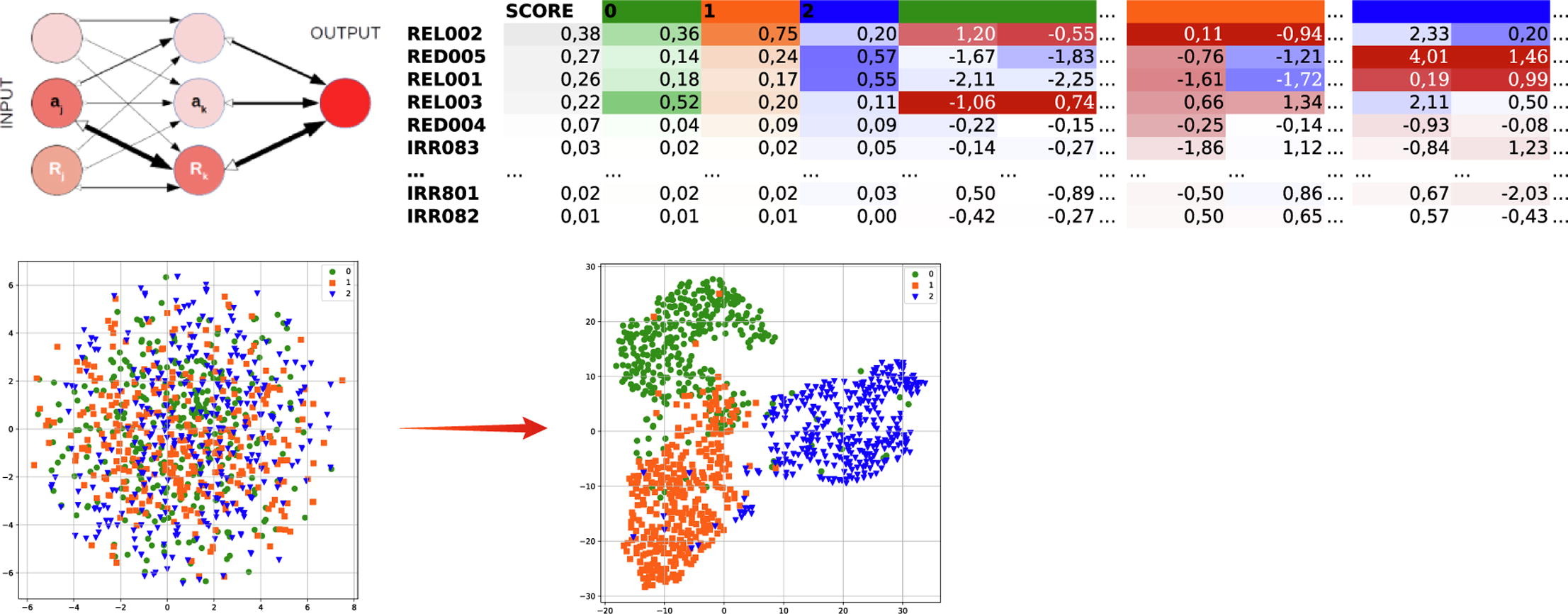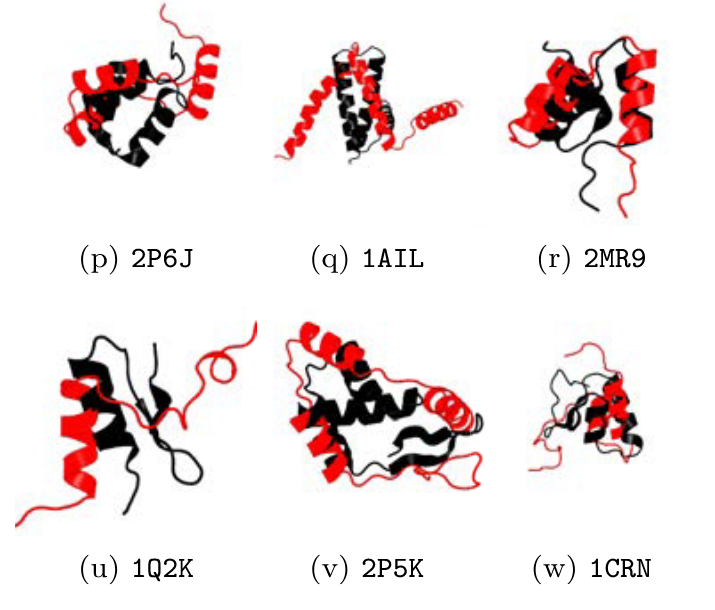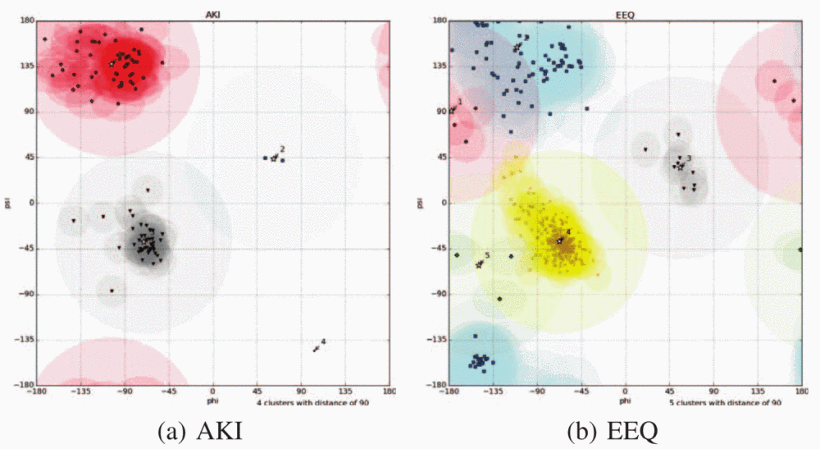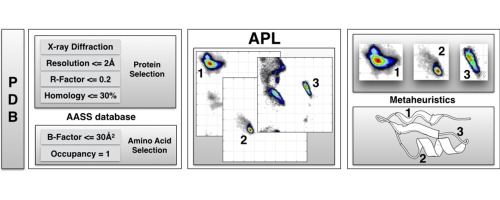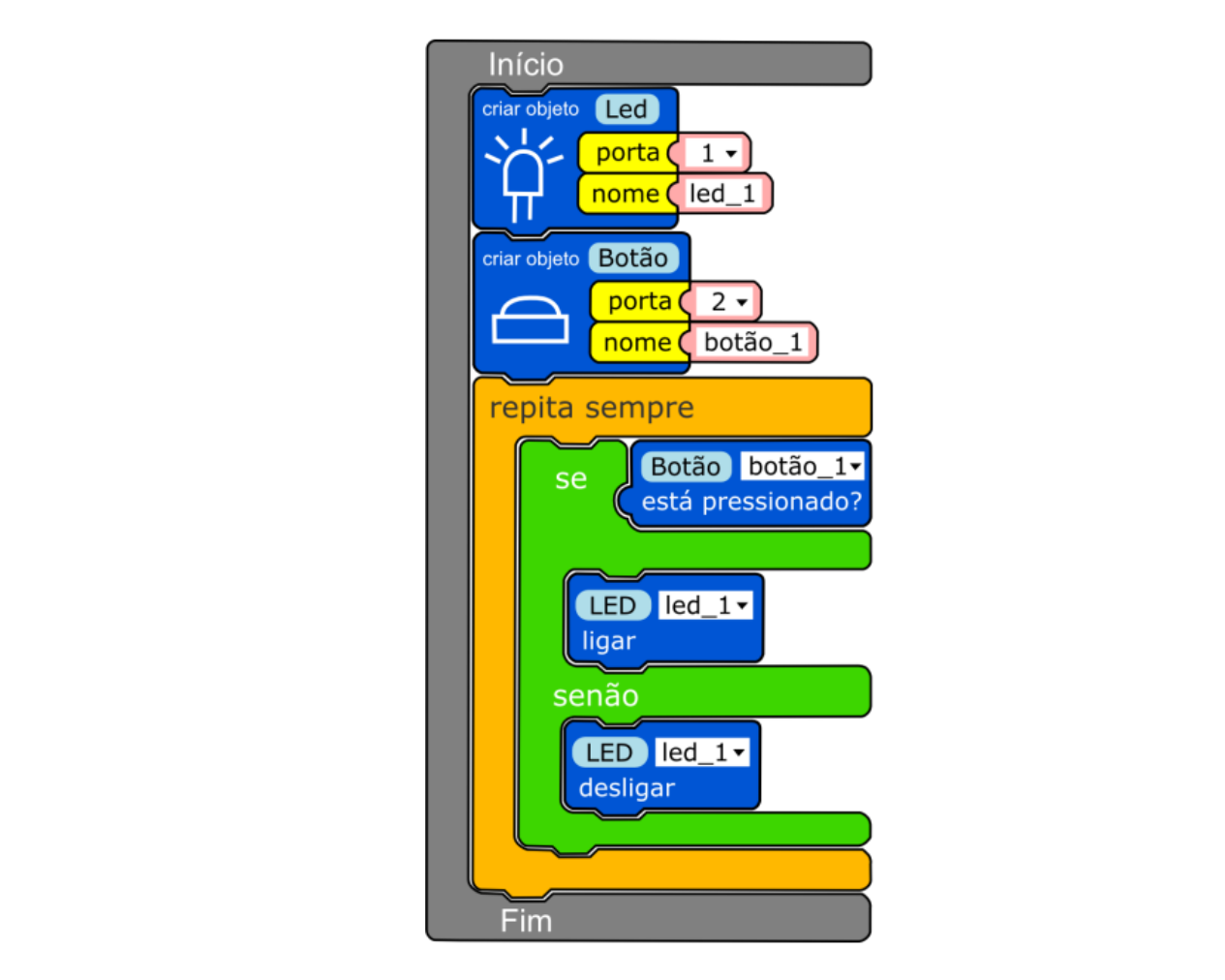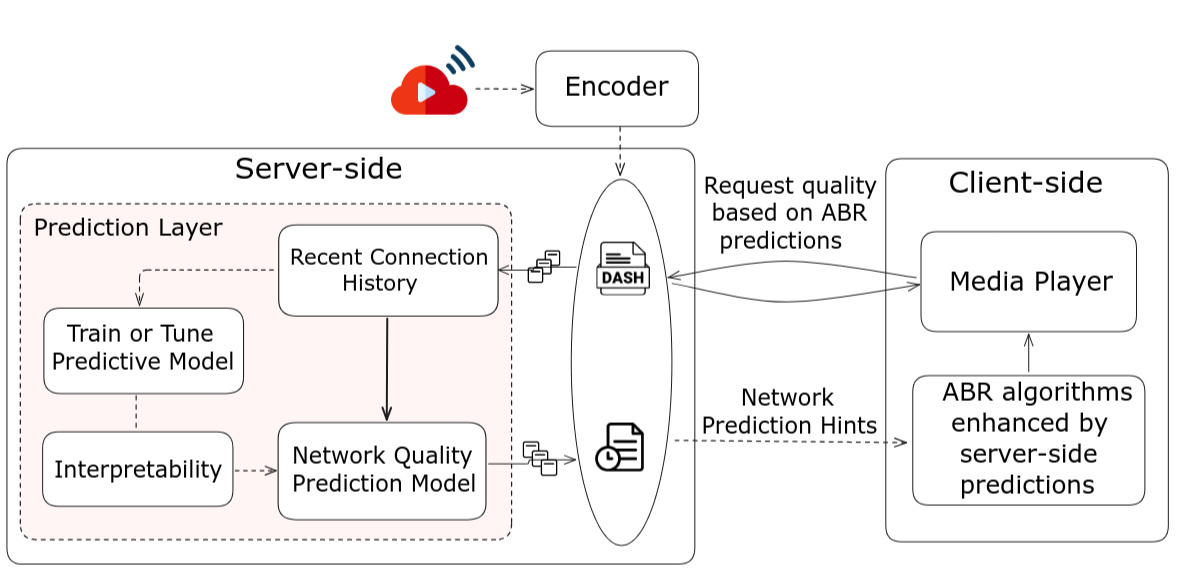
Towards an ML Assisted DASH-based Architecture: Leveraging Predictive Network Analyses with Interpretability
Eduardo R Peretto, Manoel Narciso Reis Soares Filho, Débora Cristina Santos Sousa, Luciano Paschoal Gaspary, Bruno Iochins Grisci
21st International Conference on Network and Service Management (CNSM), Bologna, Italy, 2025
📚 PDF | ResearchGate